Preparation of Solutions
Preparation of 8% DNA Sequencing Gel
| Components | Cat. No | Amount to use |
| 20% Acrylamide | ||
| Acrylamide BPE170 | 96,5g | |
| Bis-acrylamide | BPE71 | 3,35g |
| Urea | BPE169 | 233,5g |
| 5X TBE | — | 100mL |
| H2O | — | to 500mL |
| Urea Mix | ||
| Urea | BPE169 | 233,5g |
| 5X TBE | — | 100mL |
| H2O | — | to 500mL |
| 5X TBE | ||
| Tris Base | BPE152 | 54g |
| Boric acid | BPE168 | 27,5g |
| 0,5M EDTA (pH 8,0) | BPE120 | 20mL |
| H2O — | to 1L | |
| To make an 8% sequencing gel, mix the following in a small flask: | ||
| 20% Acrylamide mix | 20mL | |
| Urea mix | 30mL | |
| 10% ammonium persulfate | 0,4mL | |
| (freshly dissolved in water) | ||
For the solution into the barrel of a 50mL syringe and add 50μL of TEMED (BPE150). Mix
rapidly and inject the contents of the syringe (no needle should be used) into a preformed
sequencing gel mould.
Note: Convenient, ready made Fisher BioReagent solutions for key components are also
available. See also Tris-glycine, TBE buffers, acrylamide solutions and water.
Preparation of Tris•CI [Tris(hydroxymethyl)aminomethane] Stock Solutions (1)
Method A
- Dissolve 121g Tris Base in 800mL H2O
- Adjust solution to desired pH with concentrated HCl
- Mix and add H2O to 1L.
Method B
- Prepare a 0.1M Tris Base solution: add H20 to 12.1g Tris Base to a total volume of 1L
- From the chart below, obtain the volume of 0.1M HCl needed to produce the desired pH, and add to 100mL of 0.1M Tris Base
- Mix well
| pH, | 0,1M | pH, | 0,1M | pH, | 0,1M |
| 25°C | HCl | 25°C | HCl | 25°C | HCl |
| 7,2 | 89,4mL | 7,8 | 69,0mL | 8,4 | 34,4mL |
| 7,3 | 86,8 | 7,9 | 64 | 8,5 | 29,4 |
| 7,4 | 84 | 8 | 58,4 | 8,6 | 24,8 |
| 7,5 | 80,6 | 8,1 | 52,4 | 8,7 | 20,6 |
| 7,6 | 77 | 8,2 | 45,8 | 8,8 | 17 |
| 7,7 | 73,2 | 8,3 | 39,8 | 8,9 | 14 |
Note: The pH of Tris buffers changes significantly with temperature, decreasing approximately 0.028 pH units per 1°C. Tris-buffered solutions should be adjusted to the desired pH at the temperature at which they will be used. Since the pKa of Tris is 8.08, Tris should not be used as a buffer below pH~7.2 or above pH~9.0.
(1) Convenient, ready made Fisher BioReagent solutions are also available.
See also Tris TBE, SSC/SSPE and water
Preparation of Polyacrylamide Stacking and Separating Gels (SDS-PAGE)
| Separating gel (Total volume 15 mL)(1) | ||||||||||
| Final % acrylamide in gel(2) | 5 | 6 | 7 | 7,5 | 8 | 9 | 10 | 12 | 13 | 15 |
| Stock solutions(3) | ||||||||||
| 30% Acrylamide/0,8% Bis-acrylamide | 2,50mL | 3,00mL | 3,50mL | 3,75mL | 4,00mL | 4,50mL | 5,00mL | 6,00mL | 6,50mL | 7,50mL |
| 4X Tris Cl, pH 8,8 | 3,75 | 3,75 | 3,75 | 3,75 | 3,75 | 3,75 | 3,75 | 3,75 | 3,75 | 3,75 |
| H2O(4) 10% SDS | 8,60 0,15 | 8,10 0,15 | 7,60 0,15 | 7,35 0,15 | 7,10 0,15 | 6,60 0,15 | 6,10 0,15 | 5,10 0,15 | 4,60 0,15 | 3,60 0,15 |
| 10% Ammonium persulfate(5) | 0,05 | 0,05 | 0,05 | 0,05 | 0,05 | 0,05 | 0,05 | 0,05 | 0,05 | 0,05 |
| TEMED | 0,01 | 0,01 | 0,01 | 0,01 | 0,01 | 0,01 | 0,01 | 0,01 | 0,01 | 0,01 |
Properties of Aqueous CsCI Solutions @ 20°C
| Concentration Percent (w/w) | Concentration Percent (w/v) | Density (g/mL) | Refractive index @ 20°C |
| 20 | 23,51 | 1,1756 | 1,3507 |
| 22 | 26,33 | 1,1967 | 1,3528 |
| 24 | 29,24 | 1,2185 | 1,3550 |
| 26 | 32,27 | 1,2411 | 1,3572 |
| 28 | 35,40 | 1,2644 | 1,3594 |
| 30 | 38,66 | 1,2885 | 1,3617 |
| 32 | 42,03 | 1,3135 | 1,3641 |
| 34 | 45,54 | 1,3393 | 1,3666 |
| 36 | 49,18 | 1,3661 | 1,3691 |
| 38 | 52,96 | 1,3938 | 1,3717 |
| 40 | 56,90 | 1,4226 | 1,3744 |
| 42 | 61,00 | 1,4525 | 1,3771 |
| 44 | 65,27 | 1,4835 | 1,3800 |
| 46 | 69,73 | 1,5158 | 1,3829 |
| 48 | 74,37 | 1,5495 | 1,3860 |
| 50 | 79,23 | 1,5846 | 1,3892 |
| 52 | 84,30 | 1,6212 | 1,3925 |
| 54 | 89,62 | 1,6596 | 1,3960 |
| 56 | 95,19 | 1,6999 | 1,3996 |
| 58 | 101,05 | 1,7422 | 1,4035 |
| 60 | 107,21 | 1,7868 | 1,4076 |
| 62 | 113,71 | 1,8340 | 1,4120 |
| 64 | 120,59 | 1,8842 | 1,4167 |
Southern Blotting Stock Solutions
Prehybridisation Solution (use 0.2mL/cm2 of membrane)
- 6X SSC (BPE1325)
- 0.5% SDS (BPE1311)
- 5X Denhardt's Reagent (BPE515 or see below)
- 100μg/mL denatured salmon sperm DNA
Hybridisation Solution (use 50μL/cm2 of membrane)
- 6X SSC (BPE1325)
- 0.5% SDS (BPE1311)
- 5X Denhardt's Reagent (BPE515 or see below)
- 100μg/mL denatured salmon sperm DNA
- 0.1M EDTA
- Radioactive or non-radioactive probe DNA
Electrophoresis Stains and Tracking Dyes
| Stain/Dye | Use |
| Protein stains | |
| Silver stain | Most sensitive protein stain available |
| Coomassie* Brilliant Blue R-250 | General protein stain; more sensitive than Coomassie G-250 |
| Coomassie* Brilliant Blue G-250 | General protein stain |
| Alcian Blue | Glycoprotein stain |
| Fast Green FCF | Protein stain for collagenous tissues |
| Crocein Scarlet | Stain used in immunoelectrophoresis |
| Oil Red O | Lipoprotein stain |
| Light Green SF, Yellowish | Protein stain for collagenous tissues |
| Ponceau S | General protein stain |
| Nucleic acid stains | |
| Methylene Blue | Stain for RNA or RNase |
| Toluidine Blue O | Stain for RNA or RNase (alternative to Methylene Blue) |
| Ethidium Bromide | General nucleic acid stain |
| Silver Stain | Very sensitive nucleic acid stain for polyacrylamide gels |
| Acridine Orange | Nucleic acid stain |
| General stains (Proteins and nucleic acids) | |
| Amido Black 10B | General stain, especially suited for nitrocellulose |
| Basic Fuchsin | Glycoprotein and nucleic acid stain |
| Stains-all | General protein and nucleic acid stain |
| Tracking dyes | |
| Bromophenol Blue | Neutral and basic tracking dye, especially used for SDS–PAGE and DNA sequencing |
| Bromocresol Green | Tracking dye for DNA agarose electrophoresis |
| Pyronin Y | Acidic tracking dye for RNA electrophoresis |
| Methyl Green | Neutral and acidic tracking dye for native DNA |
| Methyl Red | Acidic tracking dye, especially used for isoelectric focusing |
| Xylene Cyanole FF | Tracking dye for DNA sequencing |
Haploid DNA Content of Various Organisms
| Size of DNA | Weight of DNA | |
| Organism | (bp) | (Daltons) |
| Mammals | ~3,0 x 109 | ~1,9 x 1012 |
| Drosophila | ~1,2 x 108 | ~7,7 x 1010 |
| Yeast (S. cerevisiae) | ~1,6 x 107 | ~1,0 x 1010 |
| E. coli | ~4,0 x 106 | ~2,5 x 109 |
| Bacteriophage T2 | ~2,0 x 105 | ~1,3 x 108 |
| Bacteriophage ë | 48,514 | 3,1 x 107 |
| pBR322 | 4363 | 2,8 x 106 |
| pUC18/pUC19 | 2686 | 1,7 x 106 |
From Molecular Cloning: A Laboratory Manual, second edition (1989).
Concentration of DNA in Solution
| Double-stranded DNA (50ìg/mL) | Molecules/mL | Moles/mL | Molar concentration | Molar concentration of termini |
| Bacteriophage | 9,78 x 1011 | 1,62 x 10-12 | 1,62nM | 3,24nM |
| pBR322 | 1,09 x 1013 | 1,81 x 10-11 | 18,1nM | 36,2nM |
| pUC18/pUC19 | 1,77 x 1013 | 2,94 x 10-11 | 29,4nM | 58,8nM |
| Segment of DNA (1kb) | 4,74 x 1013 | 7,87 x 10-11 | 78,7nM | 157,4nM |
| Octameric double-stranded linker | 5,92 x 1015 | 9,83 x 10-9 | 9,83µM | 19,7µM |
From Molecular Cloning: A Laboratory Manual, second edition (1989).
Preparation of 0.1M Potassium Phosphate Buffer @ 25°C
| Desired pH | Volume of 1M K2HPO4 (mL) | Volume of 1M KH2PO4 (mL) |
| 5,8 | 8,5 | 91,5 |
| 6,0 | 13,2 | 86,8 |
| 6,2 | 19,2 | 80,8 |
| 6,4 | 27,8 | 72,2 |
| 6,6 | 38,1 | 61,9 |
| 6,8 | 49,7 | 50,3 |
| 7,0 | 61,5 | 38,5 |
| 7,2 | 71,7 | 28,3 |
| 7,4 | 80,2 | 19,8 |
| 7,6 | 86,6 | 13,4 |
| 7,8 | 90,8 | 9,2 |
| 8,0 | 94,0 | 6,0 |
Preparation of 0.1M Sodium Phosphate Buffer @ 25°C
| Desired pH | Volume of 1M K2HPO4 (mL) | Volume of 1M KH2PO4 (mL) |
| 5.8 | 7.9 | 92.1 |
| 6.0 | 12.0 | 88.0 |
| 6.2 | 17.8 | 82.2 |
| 6.4 | 25.5 | 74.5 |
| 6.6 | 35.2 | 64.8 |
| 6.8 | 46.3 | 53.7 |
| 7.0 | 57.7 | 42.3 |
| 7.2 | 68.4 | 31.6 |
| 7.4 | 77.4 | 22.6 |
| 7.6 | 84.5 | 15.5 |
| 7.8 | 89.6 | 10.4 |
| 8.0 | 93.2 | 6.8 |
Antibiotic Solutions
| Stock Solution (1) | Working concentration | |||
| Concentration temperature | Storage plasmids | For stringent plasmids | For relaxed | |
| Ampicillin | 50mg/mL in H2O | –20°C | 20µg/mL | 60µg/mL |
| Carbenicillin | 50mg/mL in H2O | –20°C | 20µg/mL | 60µg/mL |
| Chloramphenicol | 34mg/mL in ethanol | –20°C | 25µg/mL | 170µg/mL |
| Kanamycin | 10mg/mL in H2O | –20°C | 10µg/mL | 50µg/mL |
| Tetracycline (2) | 5mg/mL in ethanol | –20°C | 10µg/mL | 50µg/mL |
(1) Stock solutions of antibiotics dissolved in H2O should be sterilised by filtration through a 0.22μm filter. Antibiotics dissolved in ethanol need not be sterilised. Store solutions in light-tight containers.
(2) Magnesium ions are antagonists of tetracycline. Use media without magnesium salts (eg, LB medium) for selection of bacteria resistant to tetracycline.
From Molecular Cloning: A Laboratory Manual, second edition (1989).
Double-stranded DNA
Density (P) = (0.998)[G + C] + 1.660g/cm3 mole
| Plasmid DNA | 50% G + C | Density |
| RFI (supercoiled) | ds DNA | 1,709g/mL |
| RFII (Nicked) | ds DNA | 1,54g/mL |
| ss DNA | 1,726g/mL | |
| ss RNA | 1,90g/mL |
Notes
• Nicked DNA binds more Ethidium bromide than does supercoiled DNA
• Ethidium bromide at saturation decreases density of dsDNA ~0.15g/mL
• More Ethidium bromide bound = greater reduction in density
• By comparison, protein has a density of 1.33g/mL
Reference
Schildkraut, C.L., Marmur, J., and Doty, P. (1962) J. Mol. Bio. 4:430-433.
Effective Range of Separation of DNAs in Agarose gels
| Amount of Agarose in gel (%[w/v]) | Effective range of separation of linear DNA molecules (kb) |
| 0,3 | 5 to 60 |
| 0,6 | 1 to 20 |
| 0,7 | 0,8 to 10 |
| 0,9 | 0,5 to 7 |
| 1,2 | 0,4 to 6 |
| 1,5 | 0,2 to 3 |
| 2,0 | 0,1 to 2 |
From Molecular Cloning: A Laboratory Manual, second edition (1989).
Effective Range of Separation of DNAs in Polyacrylamide gels
| Acrylamide (%[w/v]) (1) | Effective range of separation (bp) | Xylene Cyanol FF2 | Bromophenol Blue (2) |
| 3,5 | 1,000 to 2,000 | 460 | 100 |
| 5 | 80 to 500 | 260 | 65 |
| 8 | 60 to 400 | 160 | 45 |
| 12 | 40 to 200 | 70 | 20 |
| 15 | 25 to 150 | 60 | 15 |
| 20 | 6 to 100 | 45 | 12 |
(1) Bis-acrylamide is included at 1/30th the concentration of acrylamide
(2) Numbers are the approximate sizes of fragments of double-stranded DNA with which the dye co-migrates
From Molecular Cloning: A Laboratory Manual, second edition (1989).
Electrophoretic Blotting Buffer
| Component | Catalogue No | Amount to use | Final conc. |
| Tris base | BPE152 | 14,5g | 20mM |
| Glycine | BPE381 | 67,0g | 150mM |
| H2O | — | 4L | |
| Methanol | BPE1105 | 1,200mL Adjust pH to 8,0 | 20% |
| H2O | 6L |
From Molecular Cloning: A Laboratory Manual, second edition (1989).
Migration Rates of Marker Dyes Through Denaturing Polyacrylamide Gels
| % Polyacrylamide | Xylene Cyanol FF1 | Bromophenol Blue1 |
| 5 | 130 | 35 |
| 6 | 106 | 26 |
| 8 | 76 | 19 |
| 10 | 55 | 12 |
| 20 | 28 | 8 |
(1) Numbers are the approximate sizes of DNA (in nucleotides) with which the marker dye co-migrates.
From Molecular Cloning: A Laboratory Manual, second edition (1989).
Relative Migration of Differen Forms of Plasmid DNA on Tris-Acetate Agarose Gels (1)
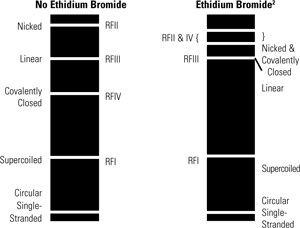
(1) The exact migration can be affected by agarose gel percentage, electrophoresis time, concentration of Ethidium Bromide, and the size and degree of supercoiling of the DNA
(2) Ethidium Bromide reduces the rate of migration of all plasmid forms. The position of covalently closed circular DNA changes relative to the other forms in the presence of Ethidium Bromide
(3) See pages 232 - 235 for Fisher BioReagents' DNA MW markers
(4) See pages 254 for Fisher BioReagents' Ethidium Bromide
From Molecular Cloning: A Laboratory Manual, second edition (1989)